Graduate Education
Weill Cornell Graduate School of Medical Sciences (WCGSMS)
For more than a half-century, WCGS has focused on preparing its students for careers in the biomedical sciences. The Graduate School faculty today numbers more than 250, and more than 1,000 students have earned Cornell University PhDs from the school.
Physiology, Biophysics and Systems Biology (PBSB)
The PBSB graduate program website includes exciting information about the research and academic activities of the Faculty engaged in world class research aiming to understand the functional mechanisms in the human body, in health and in disease.
Tri-I PhD Program in Computational Biology and Medicine (TPCBM)
The Tri-I PhD Program in Computational Biology and Medicine (CBM) was established in 2003 to provide a unique training opportunity that takes advantage of the exceptional educational and research resources of Cornell University in Ithaca, its Medical College in NYC (Weill Cornell Medical College), and Memorial Sloan Kettering Cancer Center. It is our belief that the development of such a cadre of computational biologists, trained in the laboratories of exceptional program faculty from all three campuses, will foster discovery in frontiers of basic biological and biomedical sciences.
Tri-Institutional PhD Program in Chemical Biology (TPCB)
The Tri-Institutional PhD Program in Chemical Biology was established in 2001 as one of the first graduate programs in the world to focus on research and training at the interface of chemistry and biology. The program is a collaborative offering of three premier New York City institutions, Weill Cornell Medical College, The Rockefeller University, and the Memorial Sloan Kettering Cancer Center. Located adjacent to one another in the heart of Manhattan’s Upper East Side, these three institutions combine to create a unique university environment and provide unparalleled scientific opportunities to the next generation of leaders in chemical biology.
Aksay Lab
The neural system studied by the lab is the integrator for control of eye position. Neurons essential for function, located either in the brainstem or cerebellum, are examined in vivo during normal behavior. The experimental preparation used is the developing zebrafish, allowing a combination of optical, genetic, and electrophysiological tools. Some examples of methods brought to bear are…
Elemento Lab
The Elemento lab combines Big Data analytics with experimentation to develop entirely new ways to help prevent, diagnose, understand, treat and ultimately cure cancer. Our research involves routine use of ultrafast DNA sequencing, proteomics, high-performance computing, mathematical modeling, and artificial intelligence/machine learning.
Hajirasouliha Lab
We are a computational science oriented group, affiliated with the institute of precision medicine and the institute of computational biomedicine. We are passionate about developing new algorithms and applications of computational methods to genomics.
Khelashvili Lab
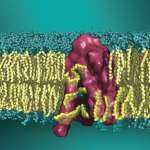
The overall goal of the research projects in the lab is to uncover dynamic mechanisms in fundamental biological processes of signal transduction by cell surface proteins in the categories of receptors (such as G protein-coupled receptors, GPCRs), transporters in the family of Neurotransmitter:Sodium-Symporters (NSS), and lipid scramblases. Special emphasis is on understanding how the spatial organization and function of these…
Khurana Lab
The research interests of the lab fall under the broad categories of genomics, computational biology and systems biology. We participate in multiple international genomics consortia and collaborate with scientists at Weill Cornell to develop novel approaches to understand the role of sequence variants in human disease. The decreasing costs of genome sequencing are leading to a growing repertoire of personal…
Krumsiek Lab
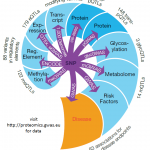
We develop innovative systems biological data analysis approaches, with a special focus on metabolomics data from human samples. By combining large datasets from modern omics profiling techniques with advanced statistics and pathway analysis methods, we attempt to unlock the true potential of such big datasets. Our working hypothesis is that only analyzing omics datasets at the level of entire molecular pathways will provide enough statistical power and biological insights to generate novel targets…
Mason Lab
Our laboratory develops and deploys computational and experimental methodologies to identify the functional genetic elements of the human genome and metagenome. To do this, we perform research in three principal areas: (1) molecular profiling in patients with extreme phenotypes, including brain malformations, aggressive cancers, and astronauts, (2) creating new biochemical and computational techniques in DNA/RNA sequencing and DNA/RNA base modifications, and (3) the development of bioinformatics models for systems…
Nirenberg Lab
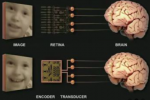
The theme of our research is to advance basic understanding of computational neuroscience, and, in parallel, use what we learn to address practical problems that improve quality of life.
Silver Lab
Our lab is focused on understanding the contributions of mast cells to a variety of diseases including bronchopulmonary dysplasia, lung and kidney fibrosis and abnormal wound healing. Mechanistically we study how mast cell mediators and exosomes communicate with cells found in close proximity, like fibroblasts. Taking a multifactorial approach we perform in vivo and in vitro experiments using murine models of…
Suhre Lab
My current research efforts are to realize the true potential of the many recent discoveries I made in the field of metabolomics and genomics, by now translating them to clinical and biomedical application. For this purpose I am involved in setting up and supporting industry-level metabolomics and proteomics facilities in Qatar, conducting clinical studies with metabolomics read-outs, developing cell-culture based…
Weinstein Lab
The Weinstein lab studies complex systems in physiology with methods of molecular and computational biophysics, bioinformatics and mathematical models. The work addresses structural and dynamic mechanisms in fundamental biological processes such as signal transduction, neuronal signaling and regulation of cell growth mechanisms, and the expression of these processes in the physiological functions of tissues and organs.
Zhu Lab
Our goal is to develop innovative multimodal single-cell genomics tools to study the combined effects of multiple regulatory layers in cell fate specification and maintenance, with an emphasis on understanding the molecular logics in development, diseases, and cancer.