
Olga Boudker, Ph.D.
- Interim Chairman of the Department of Physiology and Biophysics
- Howard Hughes Medical Institute Investigator
Whitney Pavillion
525 East 68th Street, Room W-207
New York, NY 10065
Techniques
- Bioinformatics
- Cryo-Electron Microscopy (Cryo-EM)
- Crystallography
- Förster Resonance Energy Transfer (FRET)
- Isothermal Titration Calorimetry (ITC)
- Nuclear Magnetic Resonance (NMR)
- Single-Molecule Spectroscopy
- Stopped-Flow
Research Areas
Research Summary:
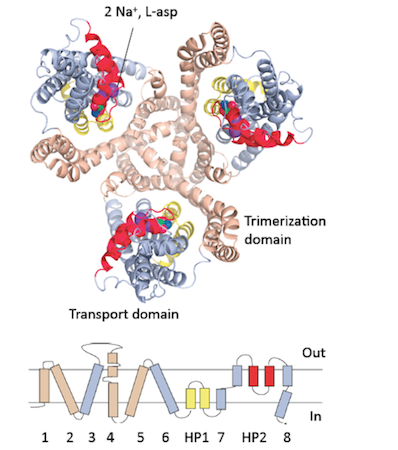
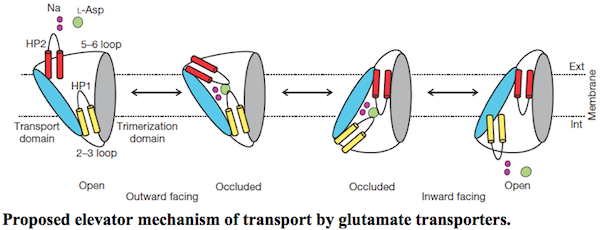
Lipid membranes define living cells, separating the internal cellular processes from the external environment. Our lab focuses on how the cells interact with the environment, especially how they move materials across the membranes. Specialized integral membrane proteins, called transporters, accomplish the task. They work as miniature machines that use the energy of ATP or trans-membrane gradients of ions to pump nutrients, neurotransmitters, waste and drugs in or out of the cells. We aim to define the molecular motions that underlie their activity. Furthermore, we decipher the structural features that define the rate at which these machines work and how they are regulated in the cells and through the use of pharmacological tools. Finally, we are interested in the evolution of transporters. Specifically, in how they have adapted to the changes of cellular environments and diversified their functional capabilities. To answer these questions we use a combination of techniques. We use crystallography and Cryo-EM to define the high-resolution structures of key functional states of transporters; single molecule FRET TIRF microscopy and NMR to probe their dynamics; biochemical approaches and isothermal titration calorimetry to probe their function and energetics; and bioinformatics to follow evolution.
Recent Publications:
- Qiu, B, Boudker, O. Structural basis of excitatory amino acid transporter 3 substrate recognition. Proc Natl Acad Sci U S A. 2025;122 (16):e2501627122. doi: 10.1073/pnas.2501627122. PubMed PMID:40249774 PubMed Central PMC12036983.
- Wang, X, Rusinova, R, Gregorio, GG, Boudker, O. Free fatty acids inhibit an ion-coupled membrane transporter by dissipating the ion gradient. J Biol Chem. 2024;300 (12):107955. doi: 10.1016/j.jbc.2024.107955. PubMed PMID:39491650 PubMed Central PMC11667161.
- Qiu, B, Boudker, O. Structural basis of the excitatory amino acid transporter 3 substrate recognition. bioRxiv. 2024; :. doi: 10.1101/2024.09.05.611541. PubMed PMID:39282329 PubMed Central PMC11398500.
- Jiang, Y, Miyagi, A, Wang, X, Qiu, B, Boudker, O, Scheuring, S et al.. HS-AFM single-molecule structural biology uncovers basis of transporter wanderlust kinetics. Nat Struct Mol Biol. 2024;31 (8):1286-1295. doi: 10.1038/s41594-024-01260-3. PubMed PMID:38632360 PubMed Central PMC11490224.
- Drew, D, Boudker, O. Ion and lipid orchestration of secondary active transport. Nature. 2024;626 (8001):963-974. doi: 10.1038/s41586-024-07062-3. PubMed PMID:38418916 .
- Fortea, E, Lee, S, Chadda, R, Argyros, Y, Sandal, P, Mahoney-Kruszka, R et al.. Author Correction: Structural basis of pH-dependent activation in a CLC transporter. Nat Struct Mol Biol. 2024;31 (4):728. doi: 10.1038/s41594-024-01242-5. PubMed PMID:38355726 .
- Fortea, E, Lee, S, Chadda, R, Argyros, Y, Sandal, P, Mahoney-Kruszka, R et al.. Structural basis of pH-dependent activation in a CLC transporter. Nat Struct Mol Biol. 2024;31 (4):644-656. doi: 10.1038/s41594-023-01210-5. PubMed PMID:38279055 PubMed Central PMC11262703.
- Reddy, KD, Rasool, B, Akher, FB, Kutlešić, N, Pant, S, Boudker, O et al.. Evolutionary analysis reveals the origin of sodium coupling in glutamate transporters. bioRxiv. 2024; :. doi: 10.1101/2023.12.03.569786. PubMed PMID:38106174 PubMed Central PMC10723334.
- Qiu, B, Boudker, O. Symport and antiport mechanisms of human glutamate transporters. Nat Commun. 2023;14 (1):2579. doi: 10.1038/s41467-023-38120-5. PubMed PMID:37142617 PubMed Central PMC10160106.
- Huang, Y, Reddy, KD, Bracken, C, Qiu, B, Zhan, W, Eliezer, D et al.. Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM. J Am Chem Soc. 2023;145 (15):8583-8592. doi: 10.1021/jacs.3c01003. PubMed PMID:37023263 PubMed Central PMC10119980.
- Reddy, KD, Ciftci, D, Scopelliti, AJ, Boudker, O. The archaeal glutamate transporter homologue GltPh shows heterogeneous substrate binding. J Gen Physiol. 2022;154 (5):. doi: 10.1085/jgp.202213131. PubMed PMID:35452090 PubMed Central PMC9044058.
- Ciftci, D, Martens, C, Ghani, VG, Blanchard, SC, Politis, A, Huysmans, GHM et al.. Linking function to global and local dynamics in an elevator-type transporter. Proc Natl Acad Sci U S A. 2021;118 (49):. doi: 10.1073/pnas.2025520118. PubMed PMID:34873050 PubMed Central PMC8670510.
- Ciftci, D, Huysmans, GHM, Wang, X, He, C, Terry, D, Zhou, Z et al.. FRET-based Microscopy Assay to Measure Activity of Membrane Amino Acid Transporters with Single-transporter Resolution. Bio Protoc. 2021;11 (7):e3970. doi: 10.21769/BioProtoc.3970. PubMed PMID:33889664 PubMed Central PMC8054209.
- Qiu, B, Matthies, D, Fortea, E, Yu, Z, Boudker, O. Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport. Sci Adv. 2021;7 (10):. doi: 10.1126/sciadv.abf5814. PubMed PMID:33658209 PubMed Central PMC7929514.
- Huysmans, GHM, Ciftci, D, Wang, X, Blanchard, SC, Boudker, O. The high-energy transition state of the glutamate transporter homologue GltPh. EMBO J. 2021;40 (1):e105415. doi: 10.15252/embj.2020105415. PubMed PMID:33185289 PubMed Central PMC7780239.
- Wang, X, Boudker, O. Large domain movements through the lipid bilayer mediate substrate release and inhibition of glutamate transporters. Elife. 2020;9 :. doi: 10.7554/eLife.58417. PubMed PMID:33155546 PubMed Central PMC7682989.
- Matin, TR, Heath, GR, Huysmans, GHM, Boudker, O, Scheuring, S. Millisecond dynamics of an unlabeled amino acid transporter. Nat Commun. 2020;11 (1):5016. doi: 10.1038/s41467-020-18811-z. PubMed PMID:33024106 PubMed Central PMC7538599.
- Ciftci, D, Huysmans, GHM, Wang, X, He, C, Terry, D, Zhou, Z et al.. Single-molecule transport kinetics of a glutamate transporter homolog shows static disorder. Sci Adv. 2020;6 (22):eaaz1949. doi: 10.1126/sciadv.aaz1949. PubMed PMID:32523985 PubMed Central PMC7259943.
- Huang, Y, Wang, X, Lv, G, Razavi, AM, Huysmans, GHM, Weinstein, H et al.. Use of paramagnetic 19F NMR to monitor domain movement in a glutamate transporter homolog. Nat Chem Biol. 2020;16 (9):1006-1012. doi: 10.1038/s41589-020-0561-6. PubMed PMID:32514183 PubMed Central PMC7442671.
- Oh, S, Boudker, O. Kinetic mechanism of coupled binding in sodium-aspartate symporter GltPh. Elife. 2018;7 :. doi: 10.7554/eLife.37291. PubMed PMID:30255846 PubMed Central PMC6175574.

