
George Khelashvili, Ph.D.
- Associate Professor of Computational Biophysics in the Institute for Computational Biomedicine
1300 York Avenue, Room LC-501A
New York, NY 10065
Techniques
Research Areas
Research Summary:
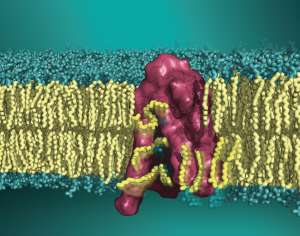
The overall goal of the research projects in the lab is to uncover dynamic mechanisms in fundamental biological processes of signal transduction by cell surface proteins in the categories of receptors (such as G protein-coupled receptors, GPCRs), transporters in the family of Neurotransmitter:Sodium-Symporters (NSS), and lipid scramblases. Special emphasis is on understanding how the spatial organization and function of these molecular machines are regulated by the cell membrane, its components (i.e. cholesterol, various lipids), and interactions with the rich environment of the cell’s proteins. We approach these research topics with advanced quantitative methods of theoretical and computational biophysics, developed and utilized at the highest level of each specialty. We pursue interdisciplinary and multi-scale strategies that integrate biophysical theory and computation with biophysical measurements and molecular cell biology experimentation. Our approach takes advantage of an abundance of molecular level insights from experimental explorations of the function and interactions of membrane-associated signaling proteins, and interprets them in a novel quantitative multi-scale framework to yield insights based on energetics, and experimentally testable hypotheses we validate with respect to mechanisms by which membrane properties and remodeling (e.g. curvature, lipid segregation) affect protein function, organization and signaling-associated interactions that are of major importance to cell physiology.

