Research topics
Mechanisms of lipid scrambling by GPCR proteins
-
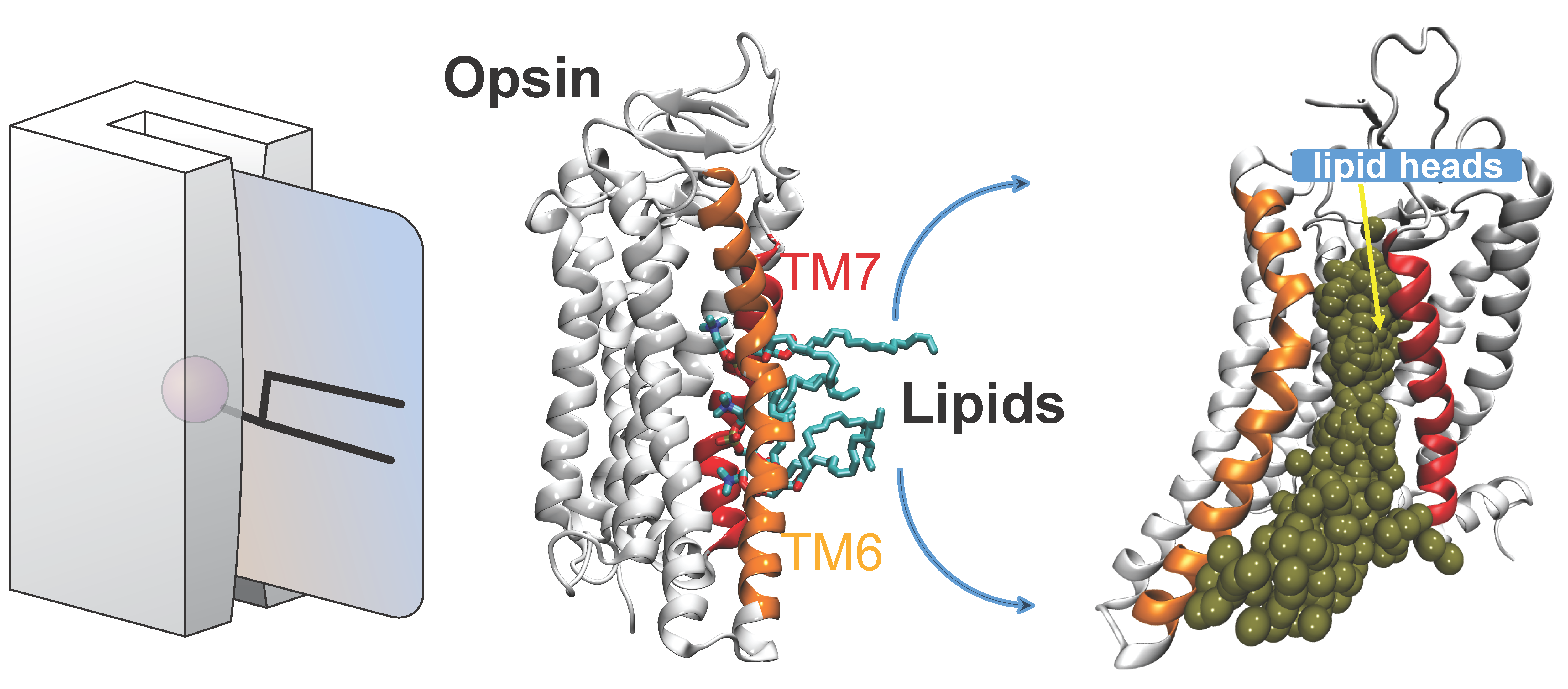
Several Class-A G protein-coupled receptor (GPCR) proteins act as constitutive phospholipid scramblases catalyzing the transbilayer translocation of >10,000 phospholipids per second when reconstituted into synthetic vesicles. To address the molecular mechanism by which these proteins facilitate rapid lipid scrambling, we carry out large-scale ensemble atomistic molecular dynamics simulations of GPCRs embedded in membranes of various lipid composition. On the example of opsin, prototypical class-A GPCR, we discovered that in the process of scrambling, lipid headgroups traverse a dynamically revealed hydrophilic pathway in the region between transmembrane helices 6 and 7 of the protein while their hydrophobic tails remain in the bilayer environment. This is reminiscent of a credit card (the magnetic strip of the card corresponds to the lipid headgroup, while the body of the card represents lipid acyl chains) swiping through a credit card reader (protein). We build quantitative kinetic models of the translocation process based on Markov State Model analysis and find that key residues on the lipid translocation pathway are conserved within the Class-A GPCR family. These results illuminate new aspects of GPCR structure and dynamics while providing a rigorous basis for the design of variants of these proteins with defined scramblase activity.
Publications:
Morra G, Razavi A, Menon AK, Khelashvili G. Cholesterol populates the lipid translocation pathway to block phospholipid scrambling by a G protein-coupled receptor. Structure 2022, 30(8):1208-1217. PMID 35660161
Morra G, Razavi AM, Pandey K, Weinstein H, Menon AK, Khelashvili G. Mechanisms of Lipid Scrambling by the G Protein-Coupled Receptor Opsin. Structure 2018, 26:1-12 PMID 29290486
Ellucidating lipid scrambling and ion channel activities of TMEM16 family proteins
-
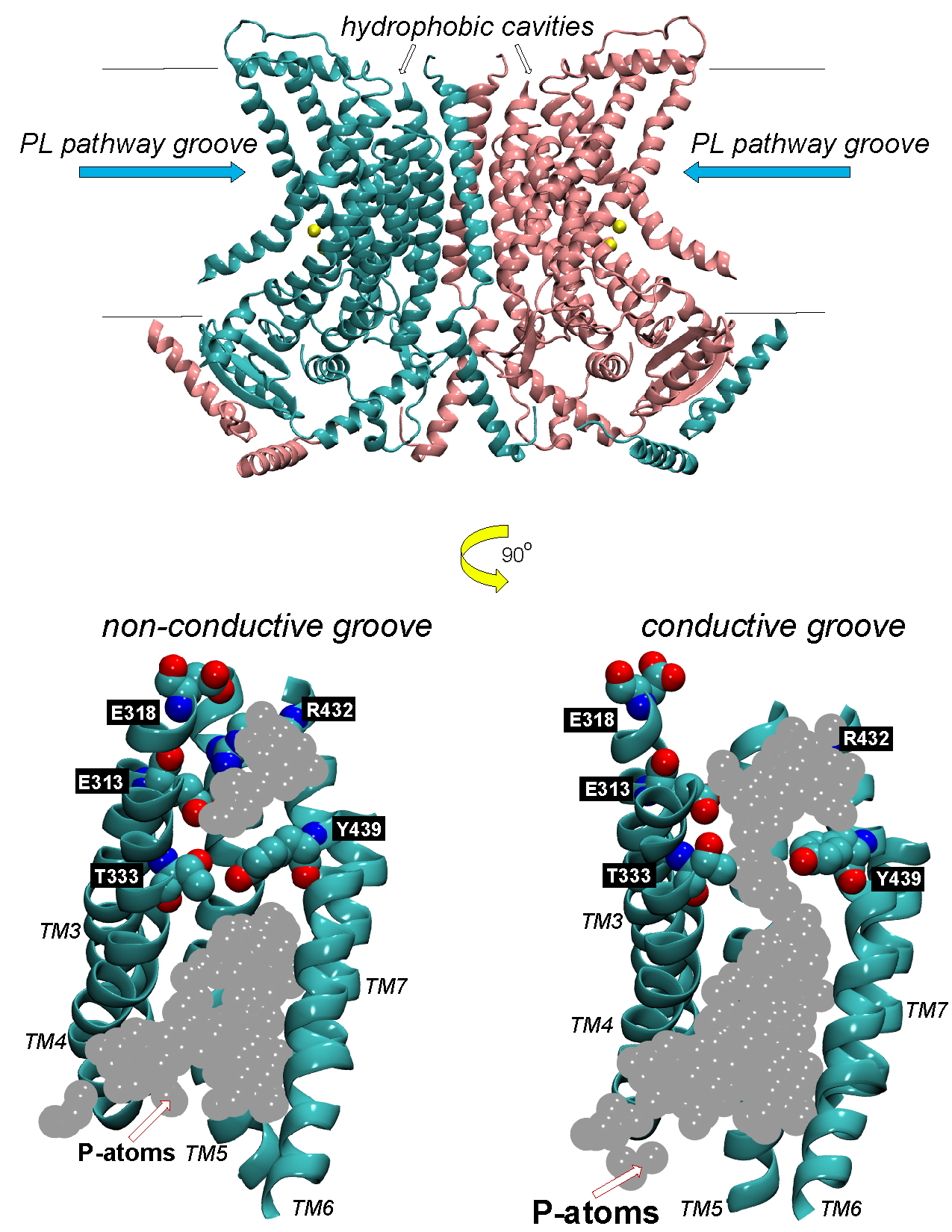
Ion transport and phospholipid (PL) scrambling are key physiological functions of membrane proteins in the TMEM16 family. The first two functionally characterized family members, TMEM16A and TMEM16B, are Ca2+-activated Cl- channels with important functions of trans-epithelial ion transport in olfaction, phototransduction, smooth muscle contraction, nociception, cell proliferation and control of neuronal excitability. Other family members, such as TMEM16E, 16F, and 16K, are PL scramblases and non-selective ion channels, and their mutations lead to genetic disorders of blood, muscle, bone and brain. The diversity of organs and tissues impacted by the impaired activity of these scramblases underscores their key roles in a wide range of physiological processes. We are investigating the molecular details of the mechanisms of TMEM16 scramblases with an integrated protocol that combines functional, structural and computational experimentation. Hypotheses generated from synergistic analyses of computational experiments and of structure-function data are tested using in vitro assays on purified TMEM16 scramblases. We are specifically interested in the dual role of the membrane as a substrate and function-supporting environment for TMEM16 scramblase activity. Predictions from large-scale molecular dynamics simulations led to the discovery of a lipid-regulated functional gate in TMEM16 scramblases. We have also uncovered the molecular mechanisms underlying the structural transformation of a TMEM16-type scramblase into a non-selective ion channel. These discoveries have brought a new understanding of the way in which the common fold of TMEM16 proteins can support the dual functions of lipid scramblase and ion channel. They brought to light specific molecular mechanisms through which membrane lipids modulate directly the function and structure of TMEM16-type scramblases. The ongoing work is directed towards: i) extending the knowledge accumulated on fungal TMEM16 scramblases to elucidate scrambling mechanisms in mammalian PLs, whose structures has been recently solved experimentally; and ii) modulation of scrambling mechanisms by the lipid membrane environment through its thermodynamic and material properties.
Publications:
Khelashvili G, Kots E, Cheng X, Levine M, Weinstein H. The allosteric mechanism leading to an open-groove lipid conductive state of the TMEM16F scramblase. Nature Communications Biology 2022, 5(1):990. PMID 36123525
Cheng X, Khelashvili G, Weinstein H. The permeation of potassium ions through the lipid scrambling path of the membrane protein nhTMEM16. Frontiers in Molecular Biosciences 2022, 9:903972. PMID 35942471
Khelashvili G, Cheng X, Falzone M, Doktorova M, Accardi A, Weinstein H. Membrane Lipids Are Both the Substrates and a Mechanistically Responsive Environment of TMEM16 Scramblase Proteins. Journal of Computational Chemistry 2020, 41, 538–551. PMID 31750558
Khelashvili G, Falzone M, Cheng X, Lee BC, Accardi A, Weinstein H. Dynamic modulation of the lipid translocation groove generates a conductive ion channel in Ca2+-bound nhTMEM16. Nature Communications 2019, 10(1):4972. PMID 31672969
Lee BC, Khelashvili G, Falzone M, Menon AK, Weinstein H, Accardi A. Gating mechanism of the extracellular entry to the lipid pathway in a TMEM16 scramblase. Nature Communications 2018, 9:3251 PMID 30108217
Investigating the functional mechanisms in ω-3 fatty acid transporter MFSD2A, a potential gateway to deliver drugs to the brain
-
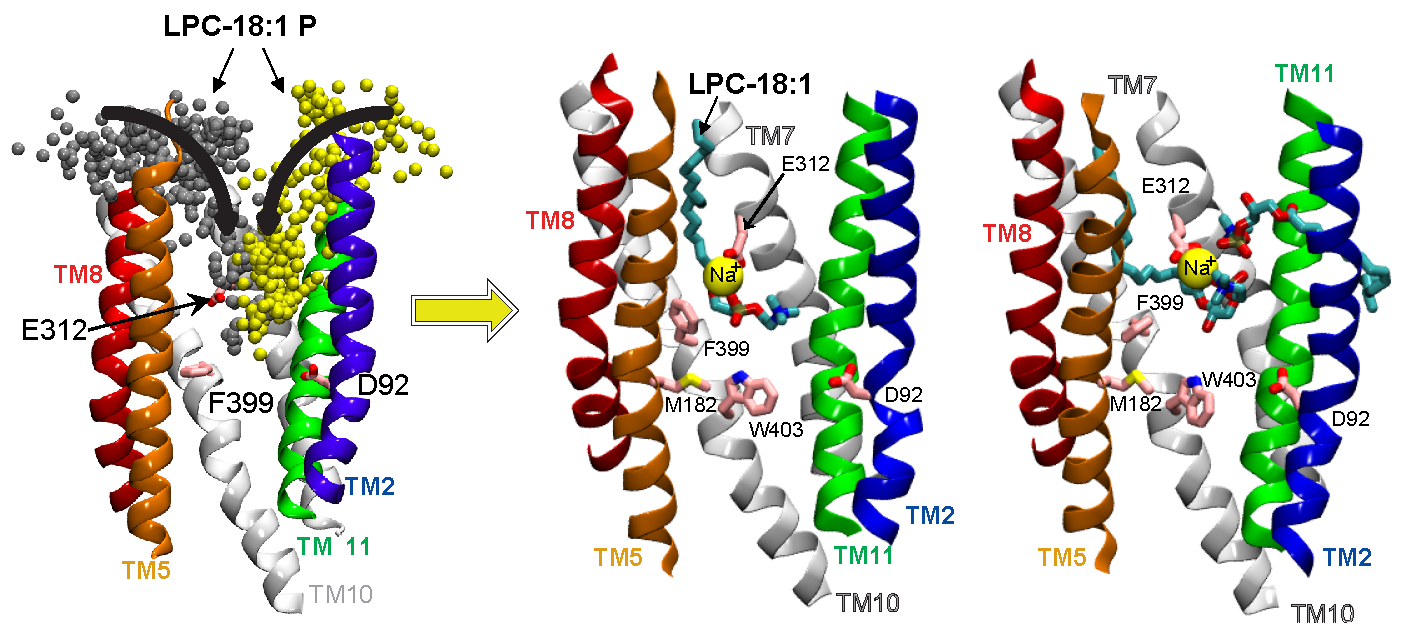
Docosahexaenoic acid (DHA) is an ω-3 fatty acid essential for neurological development and function, which is supplied to the brain and eyes predominantly from dietary sources. This nutrient is transported across the blood-brain and blood-retinal barriers in a Na+-dependent manner as lysophosphatidylcholine (LPC-DHA). This is performed by the Major Facilitator Superfamily Domain containing 2A (MFSD2A) protein. Utilizing our powerful collaborative approach of combining structure determination with single-particle cryo-electron microscopy, functional analyses, and our large-scale MD simulations, we were able to uncover for the first time the mechanism by which the MFSD2A transporter interacts with substrates, and how the Na+-dependent conformational changes release these substrates into the membrane through a lateral gate. This work provides insight into the molecular mechanism by which this atypical MFS transporter mediates uptake of single-chain phospholipids into the brain and has the potential of aiding drug delivery by guiding structure-based LPC-prodrug design for neurotherapeutic intervention. The future efforts in the lab will probe how the transport cycle in MFSD2A is mechanistically initiated allowing substrates to bind the transporter on the extracellular leaflet, which will enable understanding of mechanisms of full transport cycle in this important transporter.
Publications:
Bergman S, Cater R, Plante A, Mancia F, Khelashvili G. Substrate binding-induced conformational transitions in the omega-3 fatty acid transporter MFSD2A. Nature Communications 2023, 14(1):3391. PMID 37296098.
Cater R, Chua G-L, Erramilli S, Keener J, Choy B, Tokarz P, Chin C, Quek D, Kloss B, Pepe J, Parisi G, Wong B, Clarke O, Marty M, Kossiakoff A, Khelashvili G, Silver D, Mancia F. Structural basis of MFSD2A-mediated ω-3 fatty acid transport. Nature 2021, 595(7866):315-319. PMID 34135507
Structural basis of receptor-mediated cellular vitamin A uptake
-
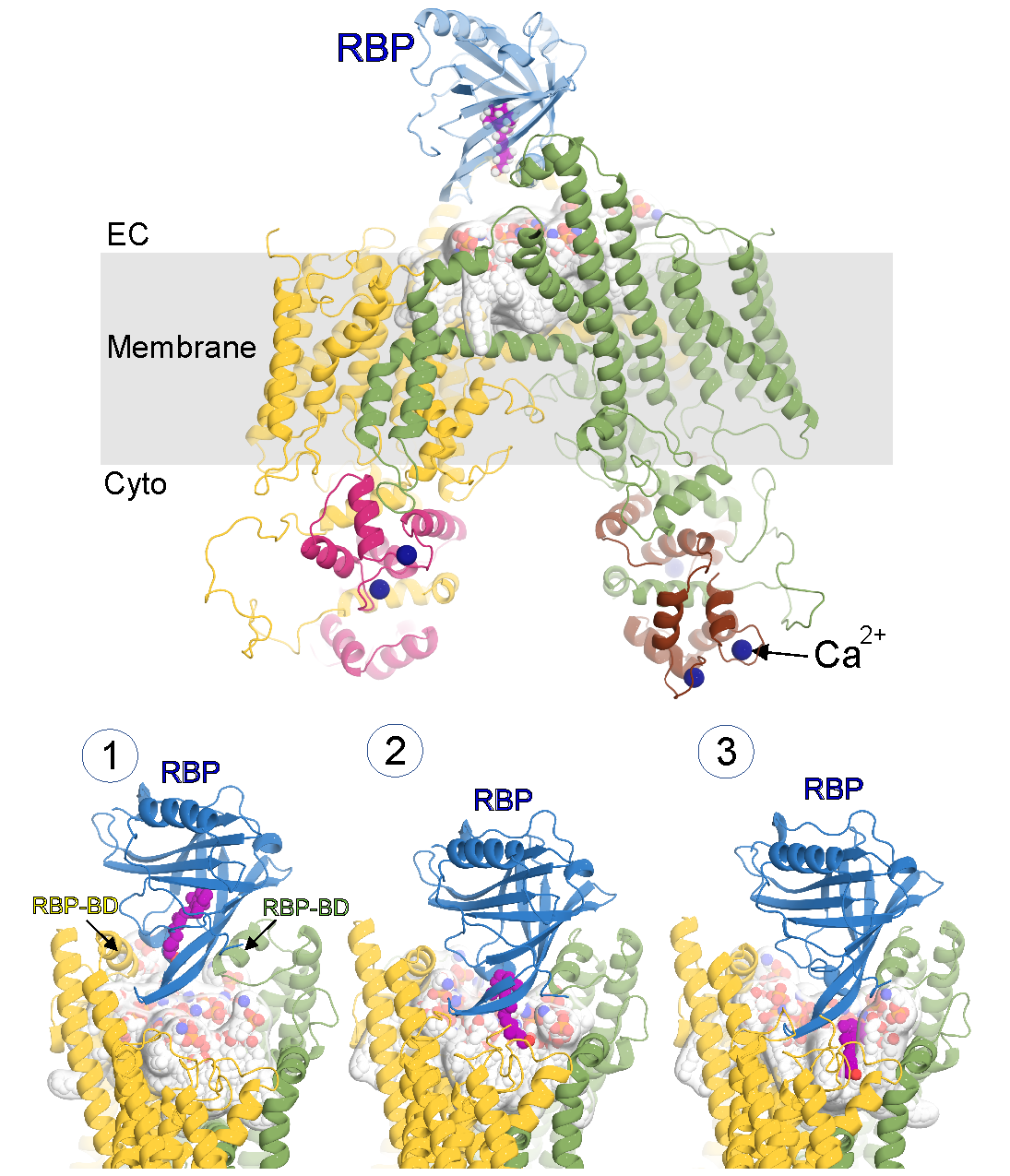
Vitamin A is an essential nutrient for all mammals and it is vital for vision. Retinol (ROH; vitamin A alcohol) is the predominant circulating form. In the fasting state, the highly hydrophobic ROH is mobilized from liver stores and solubilized by retinol-binding protein 4 (RBP). Once internalized into cells, ROH binds to cellular retinol-binding proteins (CRBPs). The RBP receptor, which proved to be encoded by a gene previously named stimulated by retinoic acid 6 (STRA6), was cloned in 2007. STRA6 is expressed widely, with particular abundance in the eye. Indeed, mutations in human STRA6 have been linked to Matthew Wood syndrome (MWS), which presents ocular defects ranging from mild microphthalmia to anophthalmia. STRA6 knockout mice exhibit defects in the visual cycle. The first structure of STRA6 determined from single-particle cryo-electron microscopy, provided a hypothesis as to how STRA6-mediated exchange of ROH from RBP might occur, via a central hydrophobic cleft into which the ROH is delivered by RBP to then diffuse into the membrane via a lateral opening. The structure also showed, unexpectedly, that STRA6 is tightly associated with calmodulin (CaM), providing an unanticipated link between retinoid metabolism and calcium homeostasis. However, several outstanding questions still remain related to i) mechanisms underlying STRA6-mediated exchange of ROH from RBP in mammals, ii) the role the membrane plays in the process, and iii) how the process may be regulated by calcium, via calmodulin. Understanding these fundamental questions will enable unprecedented insight into how the system works, at a molecular level of detail, in physiological and ultimately pathological states. To this end, we utilize an integrated approach comprising structural biology, molecular dynamics (MD) simulations and advanced statistical mechanics methods of MD data analysis, and biophysical and biochemical functional assays.
Quantification of membrane mechanical properties from molecular dynamics simulations
-
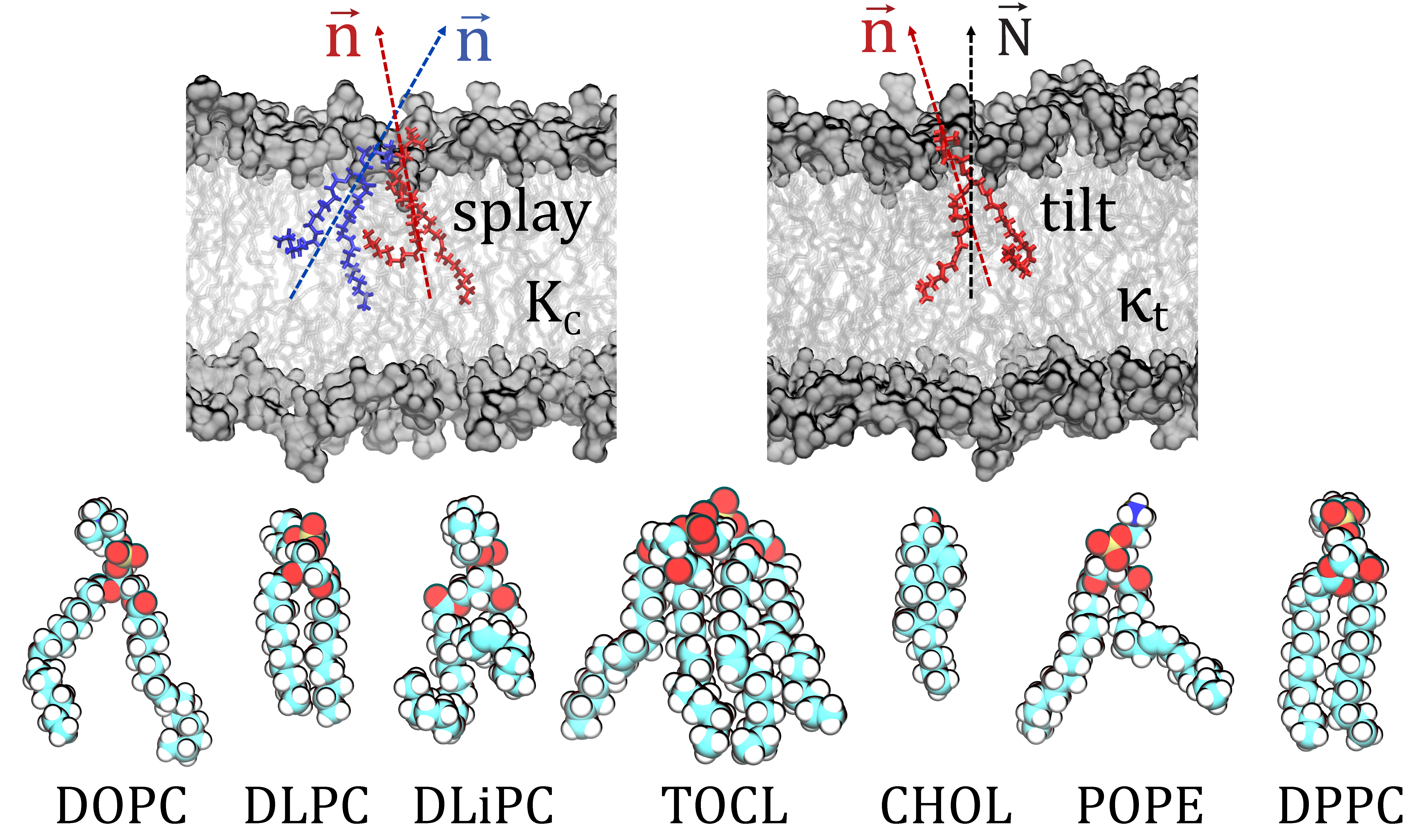
Elastic properties of membranes play major roles in many cellular processes that require membrane reshaping, including exocytosis and endocytosis. They also regulate the organization and function of many membrane-associated proteins. The determination of these properties is therefore key for our understanding of many physiological processes at the molecular level. We have developed a novel computational method, termed RSF (Real-Space Fluctuations), to quantify bending rigidity and lipid tilt modulus, the two commonly sought elastic properties of lipid assemblies. To calculate these moduli, we analyze the fluctuations in the tilt and splay degrees of freedom as sampled in the molecular dynamics trajectories in real space. The method has a number of important advantages over existing alternative computational techniques that extract bending and tilt moduli from MD simulations: i) it is not limited to single component membranes but rather is applicable to lipid mixtures; ii) it can be used to characterize lipid systems in different thermodynamic phases (i.e. liquid ordered or disorder states) and of arbitrary geometries (i.e. hexagonal phase); and iii) the local nature of the formulation allows calculations of the material properties for compositionally asymmetric and/or phase-separated lipid membrane as well as for specific membrane domains, as encountered for example in the vicinity of a protein insertion or adsorption site. Currently the work is ongoing to enhance the methodology to include calculations of other imporant components of the membrane deformation energy such as bilayer compressibility modulus.
Publications:
Chakraborty S, Doktorova M, Molugu TR, Heberle FA, Scott HL, Dzikovski B, Nagao M, Stingaciu LR, Standaert RF, Barrera F, Katsaras J, Khelashvili G, Brown MF, Ashkar R. How Cholesterol Stiffens Unsaturated Lipid Membranes. PNAS 2020, 117(36):21896-21905. PMID 32843347
Doktorova M, Harries D, Khelashvili G. Determination of bending rigidity and tilt modulus of lipid membranes from real-space fluctuation analysis of molecular dynamics simulations. Physical Chemistry Chemical Physics 2017, 19:16806-16818 PMID 28627570
Johner N, Harries D, Khelashvili G. Implementation of a methodology for determining elastic properties of lipid assemblies from molecular dynamics simulations. BMC Bioinformatics 2016, 17:161 PMID 27071656
Effect of environment on organization and function of transmembrane proteins
-

Structure/function relationships in many transmembrane proteins are often studied in artificial environments, such as detergent micelles or non-lamellar lipidic assemblies, i.e. lipid cubic phases. But how these non-native conditions influence the experimental measurements is not well understood. To evaluate the relation between the properties of the proteins measured in such arteficial media and in native conditions we study with various computational approaches structural and dynamic characteristics of different classes of transmembrane proteins (i.e. transporters, GPCRs) in detergent micelles and in lipid cubic phases.
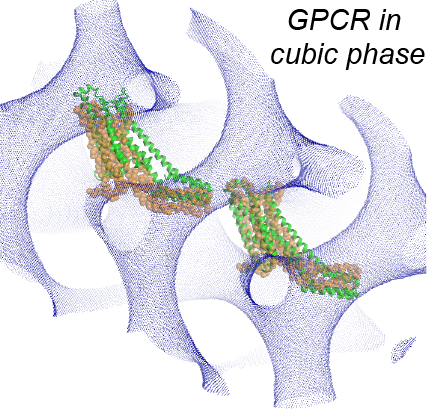
On the example of LeuT, we showed how functional properties of membrane proteins can be influenced by the identity and concentration of detergents used in the solubilization process, and demontrated a crucial role of annular lipids, that are presumably retained during the transfer of the protein from the native lipid bilayer to the detergent environment, in the experimentally measured functional phenotypes. We also showed how concentration and type of monoolein lipids can influence large scale organization of GPCR proteins in lipidic cubic phases in which these proteins are crystallized for structure determination.
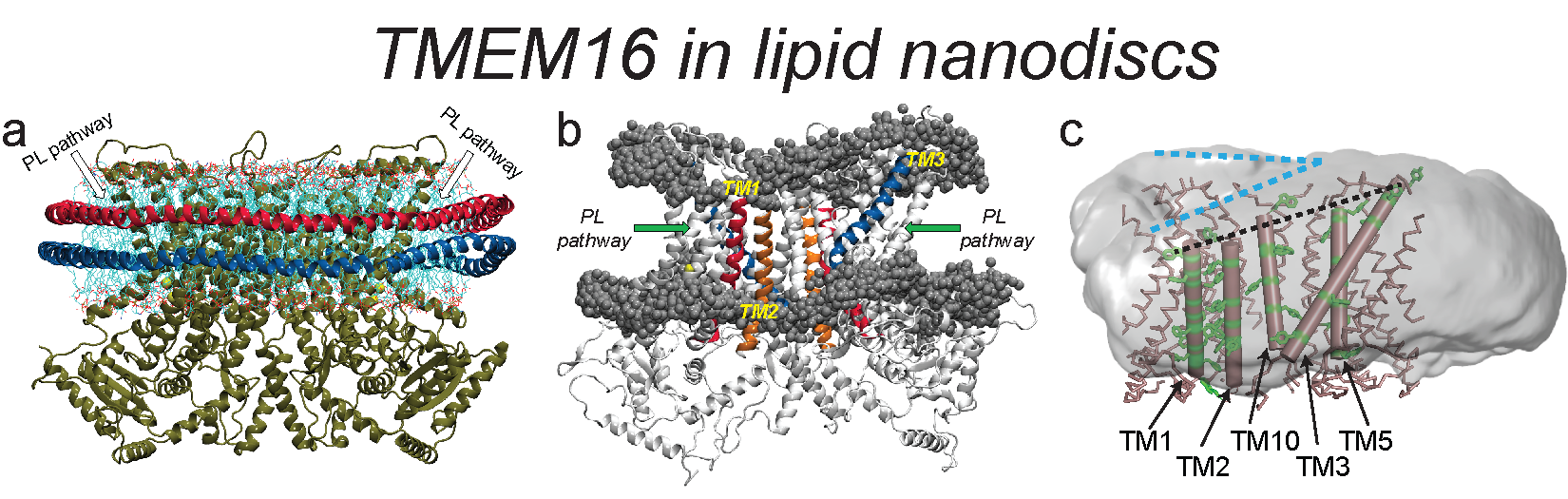
New structural information about the TMEM16 proteins is emerging from Cryo-EM determinations in lipid nanodiscs. Because the functional environment of these proteins in vivo and in in vitro is closer to flat membranes, we study comparatively the responses of the membrane to the TMEM16 proteins in flat membranes and nanodiscs. We find that bilayer shapes in the nanodiscs are very different from those observed in the flat membrane systems, but the function-related slanting of the membrane observed at the TMEM16 boundary with the membrane is similar in the nanodiscs and in the flat bilayers. This changes, however, in the bilayer composed of longer-tail lipids, which is thicker near the phospholipid translocation pathway, which may reflect an enhanced tendency of the long tails to penetrate the pathway and create, as shown previously, a non-conductive environment. These findings support the correspondence between the mechanistic involvement of the lipid environment in the flat membranes, and the nanodiscs.
Publications:
Khelashvili G, Cheng X, Falzone M, Doktorova M, Accardi A, Weinstein H. Membrane Lipids Are Both the Substrates and a Mechanistically Responsive Environment of TMEM16 Scramblase Proteins. Journal of Computational Chemistry 2020, 41, 538–551. PMID 31750558
LeVine MV, Khelashvili G, Shi L, Quick M, Javitch JA, Weinstein H. Role of Annular Lipids in the Functional Properties of Leucine Transporter LeuT Proteomicelles. Biochemistry 2016, 55:850-859 PMID 26811944
Khelashvili G, LeVine M, Shi L, Quick M, Javitch J, Weinstein H. The Membrane Protein LeuT in Micellar Systems: Aggregation Dynamics and Detergent Binding to the S2 Site. Journal of the American Chemical Society 2013 135(38):14266–14275 PMID 23980525
Johner N, Mondal S, Morra G, Caffrey M, Weinstein H, Khelashvil G. Protein and Lipid Interactions Driving Molecular Mechanisms of in meso Crystallization. Journal of the American Chemical Society 2014 136(8):3271-84 PMID 24494670
