
Ashley Laughney, Ph.D.
- Associate Professor of Computational Cancer Genomics in Computational Biomedicine in the Institute for Computational Biomedicine
1305 York Avenue, Room Y.13.04
New York, New York 10021
Techniques
- Animal Models (e.g., zebrafish)
- Bioinformatics
- Cell Sorting
- Confocal Microscopy
- Fluorescence Microscopy and FRAP
- High Performance Computing
- Immunofluorescence microscopy
- Mathematical Modeling and Simulations
Research Areas
Research Summary:
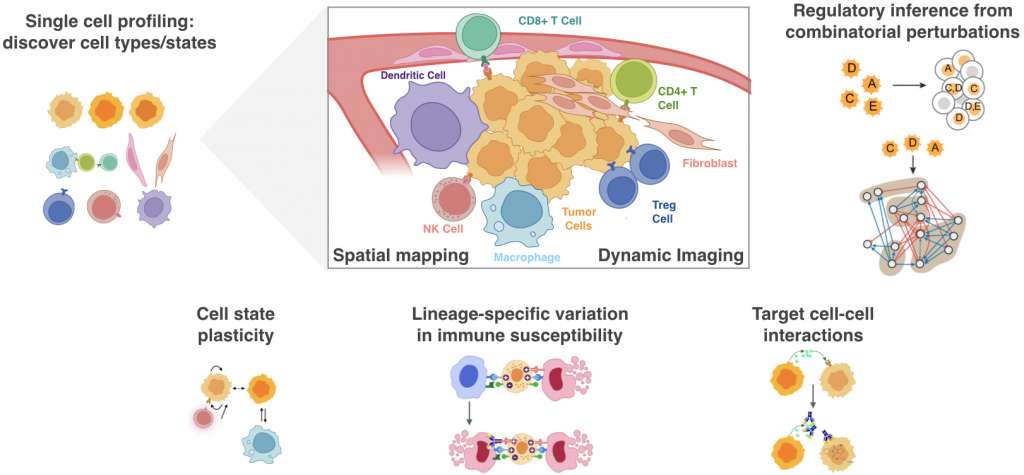
Intra-tumor heterogeneity is a hallmark of cancer that has important implications for our understanding of tumor biology and its therapeutic control. It has complicated efforts to target only one gene or one population of tumor cells without considering how diverse tumor cell types or phenotypic states contribute to adaptive behaviors like drug resistance and metastasis. The Laughney Lab integrates powerful systems biology methods – including massively parallel, single-cell RNA expression profiling with quantitative imaging technologies and mouse models of metastasis – to discover context-dependent interactions supporting the adaptive abilities of disseminated tumor cells (DTCs) and their interplay with anti-tumor immunity. We are particularly interested in dissecting tumor cell heterogeneity through the lens of tissue regeneration and repair. The understanding of cancer as a chronic, non-healing wound, has long invoked a central role for developmental pathways underlying normal tissue repair. However, the degree and fidelity of their enactment during tumor progression and under the selective pressures of immune surveillance, remains largely unknown. To address this knowledge gap, we are systematically dissecting lineage specific immune vulnerabilities across epithelial cell states observed in development and cancer progression. We also are exploring functional redundancies in key pathways that promote an immunogenic response by combining single cell methods with targeted perturbations. By integrating systems biology methods within a hypothesis driven framework, our goal is to better understand how developmental plasticity amongst individual cancer cells shapes immune susceptibility and lethal transitions like metastasis or the emergence of drug resistance.
Recent Publications:
- Di Bona, M, Chen, Y, Agustinus, AS, Mazzagatti, A, Duran, MA, Deyell, M et al.. Micronuclear collapse from oxidative damage. Science. 2024;385 (6712):eadj8691. doi: 10.1126/science.adj8691. PubMed PMID:39208110 PubMed Central PMC11610459.
- Gardner, EE, Earlie, EM, Li, K, Thomas, J, Hubisz, MJ, Stein, BD et al.. Lineage-specific intolerance to oncogenic drivers restricts histological transformation. Science. 2024;383 (6683):eadj1415. doi: 10.1126/science.adj1415. PubMed PMID:38330136 PubMed Central PMC11155264.
- Rogava, M, Aprati, TJ, Chi, WY, Melms, JC, Hug, C, Davis, SH et al.. Loss of Pip4k2c confers liver-metastatic organotropism through insulin-dependent PI3K-AKT pathway activation. Nat Cancer. 2024;5 (3):433-447. doi: 10.1038/s43018-023-00704-x. PubMed PMID:38286827 PubMed Central PMC11175596.
- Li, J, Hubisz, MJ, Earlie, EM, Duran, MA, Hong, C, Varela, AA et al.. Non-cell-autonomous cancer progression from chromosomal instability. Nature. 2023;620 (7976):1080-1088. doi: 10.1038/s41586-023-06464-z. PubMed PMID:37612508 PubMed Central PMC10468402.
- Adler, FR, Anderson, ARA, Bhushan, A, Bogdan, P, Bravo-Cordero, JJ, Brock, A et al.. Modeling collective cell behavior in cancer: Perspectives from an interdisciplinary conversation. Cell Syst. 2023;14 (4):252-257. doi: 10.1016/j.cels.2023.03.002. PubMed PMID:37080161 PubMed Central PMC10760508.
- Bhardwaj, P, Iyengar, NM, Zahid, H, Carter, KM, Byun, DJ, Choi, MH et al.. Obesity promotes breast epithelium DNA damage in women carrying a germline mutation in BRCA1 or BRCA2. Sci Transl Med. 2023;15 (684):eade1857. doi: 10.1126/scitranslmed.ade1857. PubMed PMID:36812344 PubMed Central PMC10557057.
- Li, Z, Low, V, Luga, V, Sun, J, Earlie, E, Parang, B et al.. Tumor-produced and aging-associated oncometabolite methylmalonic acid promotes cancer-associated fibroblast activation to drive metastatic progression. Nat Commun. 2022;13 (1):6239. doi: 10.1038/s41467-022-33862-0. PubMed PMID:36266345 PubMed Central PMC9584945.
- Bakhoum, MF, Francis, JH, Agustinus, A, Earlie, EM, Di Bona, M, Abramson, DH et al.. Loss of polycomb repressive complex 1 activity and chromosomal instability drive uveal melanoma progression. Nat Commun. 2021;12 (1):5402. doi: 10.1038/s41467-021-25529-z. PubMed PMID:34518527 PubMed Central PMC8438051.
- Deyell, M, Garris, CS, Laughney, AM. Cancer metastasis as a non-healing wound. Br J Cancer. 2021;124 (9):1491-1502. doi: 10.1038/s41416-021-01309-w. PubMed PMID:33731858 PubMed Central PMC8076293.
- Haynes, K, Yau, C, Bild, A, Laughney, A, Morsut, L, Yang, X et al.. How Has the COVID-19 Pandemic Changed How You Will Approach Research and Lab Work in the Future?. Cell Syst. 2020;11 (6):550-554. doi: 10.1016/j.cels.2020.11.011. PubMed PMID:33333028 PubMed Central PMC7833873.
- Xie, XP, Laks, DR, Sun, D, Poran, A, Laughney, AM, Wang, Z et al.. High-resolution mouse subventricular zone stem-cell niche transcriptome reveals features of lineage, anatomy, and aging. Proc Natl Acad Sci U S A. 2020;117 (49):31448-31458. doi: 10.1073/pnas.2014389117. PubMed PMID:33229571 PubMed Central PMC7733854.
- Ganesh, K, Basnet, H, Kaygusuz, Y, Laughney, AM, He, L, Sharma, R et al.. Author Correction: L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat Cancer. 2020;1 (11):1128. doi: 10.1038/s43018-020-00130-3. PubMed PMID:35122071 .
- Wang, R, Sharma, R, Shen, X, Laughney, AM, Funato, K, Clark, PJ et al.. Adult Human Glioblastomas Harbor Radial Glia-like Cells. Stem Cell Reports. 2020;15 (1):275-277. doi: 10.1016/j.stemcr.2020.06.002. PubMed PMID:32668221 PubMed Central PMC7363934.
- Ganesh, K, Basnet, H, Kaygusuz, Y, Laughney, AM, He, L, Sharma, R et al.. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat Cancer. 2020;1 (1):28-45. doi: 10.1038/s43018-019-0006-x. PubMed PMID:32656539 PubMed Central PMC7351134.
- Rozenblatt-Rosen, O, Regev, A, Oberdoerffer, P, Nawy, T, Hupalowska, A, Rood, JE et al.. The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell. 2020;181 (2):236-249. doi: 10.1016/j.cell.2020.03.053. PubMed PMID:32302568 PubMed Central PMC7376497.
- Laughney, AM, Hu, J, Campbell, NR, Bakhoum, SF, Setty, M, Lavallée, VP et al.. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020;26 (2):259-269. doi: 10.1038/s41591-019-0750-6. PubMed PMID:32042191 PubMed Central PMC7021003.
- Wang, R, Sharma, R, Shen, X, Laughney, AM, Funato, K, Clark, PJ et al.. Adult Human Glioblastomas Harbor Radial Glia-like Cells. Stem Cell Reports. 2020;14 (2):338-350. doi: 10.1016/j.stemcr.2020.01.007. PubMed PMID:32004492 PubMed Central PMC7014025.
- Santos, CP, Lapi, E, Martínez de Villarreal, J, Álvaro-Espinosa, L, Fernández-Barral, A, Barbáchano, A et al.. Urothelial organoids originating from Cd49fhigh mouse stem cells display Notch-dependent differentiation capacity. Nat Commun. 2019;10 (1):4407. doi: 10.1038/s41467-019-12307-1. PubMed PMID:31562298 PubMed Central PMC6764959.
- Elizalde, S, Laughney, AM, Bakhoum, SF. A Markov chain for numerical chromosomal instability in clonally expanding populations. PLoS Comput Biol. 2018;14 (9):e1006447. doi: 10.1371/journal.pcbi.1006447. PubMed PMID:30204765 PubMed Central PMC6150543.
- Bakhoum, SF, Ngo, B, Laughney, AM, Cavallo, JA, Murphy, CJ, Ly, P et al.. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553 (7689):467-472. doi: 10.1038/nature25432. PubMed PMID:29342134 PubMed Central PMC5785464.

